LLMs in Public Health - Part 1
Three ways LLM classifiers can support public health
This blog is written in a personal capacity and all opinions are my own.
This is the first in a series of posts summarising work I've helped publish at the UK Health Security Agency assessing the potential of Large Language Models (LLMs) within public health. This blog focuses on our earliest and longest running work - evaluating and deploying LLMs for domain specific classification tasks on a broad range of public health relevant free text.
Introduction
Having all lived through a global pandemic, we are very aware of some of the ways public health interventions involve the classification and extraction of messy real world information. It could be entering symptoms on a COVID-19 contact tracing form (loss of smell!), the type of contact needed for something to be considered 'close contact', whether a person is clinically vulnerable, or simply who counts as part of your household. Consistently and accurately assigning these classifications is often a core part of enabling effective public health interventions and surveillance. Therefore, the potential for LLMs to support and help scale these tasks is an important area of research within public health today.
This post goes into our initial work within the UK Health Security Agency (UKHSA) evaluating LLMs’ potential to support public health experts: (1) build new structured datasets, (2) scale existing public health interventions, and (3) improve internal processes.
For full details, please refer to the preprints below:
Evaluating Large Language Models for Public Health Classification and Extraction Tasks (2024)
1. Using LLMs to expand public health data sources
Existing public health surveillance systems are often partly constrained by the cost and expert time required to provide testing, collect relevant information (e.g contact tracing, surveys etc.), and analyse the results. One area we explore in detail, particularly in the context of gastrointestinal (GI) illness, is whether LLMs can be used to expand the range of data sources that can feed into public health surveillance.
We focused initially on foodborne gastrointestinal (GI) illness for a few reasons:
(1) Large number of undetected cases - It is estimated that only 10% of people experiencing GI illness seek medical attention, and so there could be significant benefits if LLMs and alternative data can help us understand the undetected cases.
(2) Considerable disease burden - GI illness is a significant cause of ill health in the United Kingdom (UK) and around the world. It is estimated that there are 2.4 million cases of foodborne illness in the UK every year, resulting in a cost of £9.1 billion ($11.8b).
(3) Large relevant public free text datasets - The greatest benefits to applying LLMs are often found when you can leverage their ability to process large volumes of relevant unstructured free text. Online restaurant review platforms are a particularly rich potential source of information on whether people contracted foodborne GI illness, as affected diners may want to share their experience with the management of the establishment or other prospective diners. However, the large number of reviews means manual classification by experts is impractical. This raises the potential for LLMs to perform disease surveillance which may give some insight into GI infections not reported to existing surveillance systems.
To evaluate the feasibility of this approach, we manually annotated a dataset of 3000 restaurant reviews from the Yelp Open Dataset, with the help of experts in GI illness. Each review was classified as to whether they refer to possible instances of GI illness. Then for a subset of reviews that were identified as referring to potential GI illness, we also manually annotated the symptoms and foods referenced within the review.
We tested general purpose open-weight LLMs (Llama 3.3, Mistral Large, Gemma 2, and others) using in context learning approaches, as well as fine tuning smaller models (RoBERTa-large) specifically for this classification task. We found the highest performing models achieve over 90% Micro-F1 scores on each of the three tasks (see below). In addition, we provide preliminary bias assessments using Social Group Substitution prompting approaches.
Combined this work and the automated evaluations underpinning it provide us with a strong basis for potentially supporting real world GI illness surveillance with LLMs in the future.
This is just one example of the novel structured public health datasets that LLMs could be used to create from available free text.
2. Using LLMs to help scale existing interventions during outbreaks
A key challenge within public health, as found in the COVID-19 pandemic, is how to quickly scale interventions and surveillance during an outbreak response. Therefore, the ability of LLMs to automatically process very large volumes of free text in a scalable way could make them a useful tool when faced with rapidly growing incidents.
To understand and quantify the performance of LLMs on these outbreak response tasks, we used the 2022 mpox outbreak as a case study to evaluate the ability of LLMs to perform contact type classification - one of the common types of contact tracing classifications conducted during outbreaks. We chose the mpox outbreak for a few reasons:
(1) Novel characteristics - The 2022 mpox clade II outbreak was both the largest recent global outbreak of mpox, and in the UK cases were mainly identified in gay, bisexual, and other men who have sex with men without documented history of travel to endemic countries. These novel characteristics made this mpox outbreak a more challenging example of contact tracing.
(2) Sexual references - Text within mpox contact tracing forms often contains discussion of sexual activity, which is an important risk factor for some infections. However, many LLM pre-training and fine-tuning datasets are designed to avoid text about sexual activity or to encourage refusals when these topics arise. Therefore, evaluating performance on this type of free text is essential if using LLMs for certain disease areas in public health.
(3) Bespoke protocol - The definitions and protocol for identifying the different categories of contact observed during the mpox response were set by experts specifically for the outbreak. Therefore, this task requires the LLM to apply a detailed protocol provided within the prompt, rather than drawing on existing knowledge.
To evaluate LLMs, first experts with access to real world contact tracing responses generated a synthetic (fictional) dataset designed to replicate the type, format, and content of the real responses. This dataset was then further augmented with additional variations of the original synthetic data generated by an LLM, which were manually reviewed for errors.
We assessed open-weight LLMs using both zero-shot and few-shot prompting. Interestingly, likely due to the complexity of the protocol, we found LLMs struggled in the zero-shot (no examples) setting with the highest performing model achieving only 53% Micro-F1 score. However, with a detailed 10-shot prompt giving examples of the protocol being applied, we saw large improvements across almost all models with the highest performing nearing 80% Micro-F1 score.
Therefore, whilst the LLMs we assessed found mpox contact type classification challenging, there are promising signs that with newer models, chain of thought prompting (or reasoning), and domain specific fine tuning, LLMs may well be useful tools to support experts in future outbreaks. This could in turn support scaling contact tracing in response to rapidly growing disease outbreaks longer term.
3. Using LLMs to support existing processes
Outside of outbreaks, regular public health processes and reporting often involve large volumes of free text that currently require manual work by experts to process. LLMs are one tool that could help experts speed up or augment these existing manual workflows.
To evaluate LLMs for these types of internal task, we identified two internal processes that were particularly amenable to these approaches:
(1) Country disambiguation for TB screening - Different pathogens are endemic to different regions of the world. As such, understanding the risk profile of an individual often requires understanding their recent travel history or previous countries where they have lived. One example of this is for Tuberculosis screening, where currently experts often have to manually identify the place of birth in thousands of records per year where the location written in the free text field cannot be identified using existing automated matching.
To assess LLMs’ ability to augment this process, we use a manually annotated dataset of anonymised free text responses from GP registration form place of birth fields that could not be matched. We then evaluate LLMs' accuracy at identifying the disambiguated country. Work is ongoing to deploy this system to support TB screening.
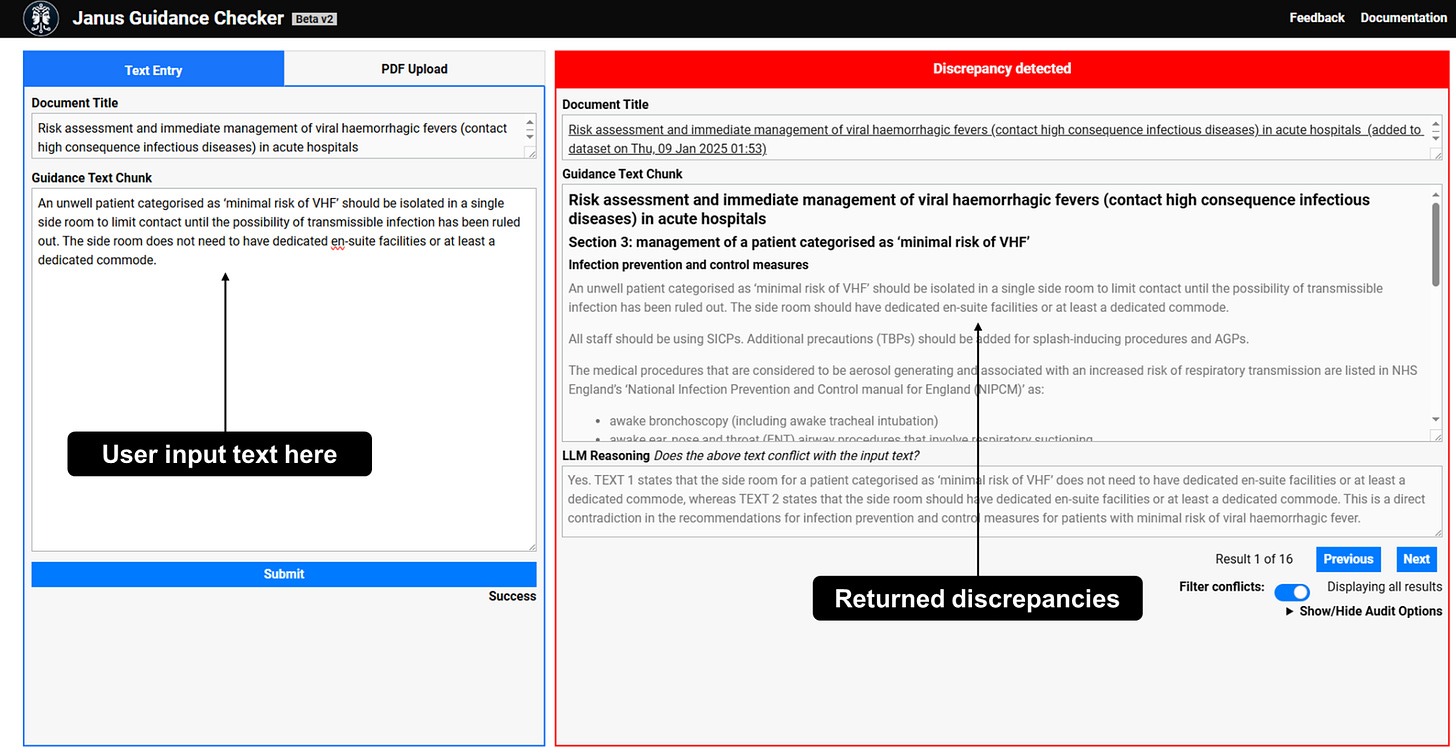
(2) Reviewing public health guidance - Providing guidance is a key role of public health institutions, and UKHSA currently provides hundreds of guidance documents for professionals and the public. However, ensuring guidance is consistent, accurate, and up to date is a continual challenge, especially in the face of rapidly changing outbreaks and incident responses.
To assess LLMs’ ability to help experts identify potential conflicts between public health guidance recommendations contained in guidance documents, we evaluated LLMs’ capacity to identify passages containing recommendations, and then detect discrepancies between pairs of passages.
We are currently developing this into a system that lets internal users upload a piece of guidance in development and automatically retrieves existing, relevant sections of UKHSA guidance, then flags any potential conflicts between them.
These are two real world examples of how LLMs are already starting to augment existing public health processes.
Conclusion
As illustrated across these evaluations, the diversity of topics, tasks, text, and incidents, means public health free text processing is a particularly challenging area. However, LLMs' broad knowledge, in-context learning, and capacity for automated processing at scale, means they could be a powerful tool to help experts perform core public health activities. This could in turn also help us respond more effectively to future pandemics. As in most domains, the first step to successful deployment is developing robust evaluations to identify tractable tasks and help incorporate LLMs safely into existing processes.
Acknowledgements
I’d like to thank all my colleagues in Advanced Analytics, Technology, and the Chief Medical Advisor group at UKHSA for helping deliver and supporting the work this blog is based on.